Towards implementing Hadoop/Spark-based next-generation NDB that stores health insurance claims data of all residents in Japan.
SERVICES / Big Data Analytics
Customer's Issues
- Rigid system structure, which is a characteristic of relational databases (RDB)
- Various rework required at each step of data use
- Cumbersome data conversion work required to reorganize the raw data into data structures suitable for various researches
Solutions
- Realization of high performance and scalability by utilizing Hadoop/Spark
- Improvement of the learning efficiency by establishing "materials to learn the basics of health insurance claims" and "a learning environment where dummy data is available"
- Increase of the analysis efficiency and convenience by preparing data marts required for analysis
Research that aims to visualize medical activities through the nationwide analysis of health insurance claims data has the potential of bringing an extensive reform to future medical services in Japan. The nationwide health insurance claims data form very large data sets with approximately 10 billion records. However, the current database (NDB) that stores health insurance claims data and specific health checkups data, has not been easy for users such as researchers and data analysts to use due to various reasons including architectural issues. This made it difficult to analyze and visualize health insurance claims data.
A project named "Research for Infrastructure Construction of Studies with the Next Generation NDB Data to Create New Evidence " brought together specialists in various fields to build the next-generation-NDB data research platform. This project has succeeded in building a high-performance and scalable system by adopting Apache Hadoop/ Apache Spark (Hadoop/Spark hereafter), which can handle unstructured data in a scalable manner, as the core technology.

Background and issues Challenges that led to the replacement of the current NDB
Major issues with current NDB — Rigid system structure and difficulty in using data
In General, health insurance unions/companies bear all or part of the medical expenses arising from medical services. In Japan, all residents are required to join the public health insurance system by law and it is said to be the best in the world. On the other hand, in the United States, where a public universal health insurance system is not available, residents have to subscribe to private health insurance policies except for elderly people, disabled people, or low income earners, etc. However, there are large number of residents in the United States without subscribing to a private health insurance policy. Although reforms for the health insurance system in the United States is under way, there are many issues. For example, medical fees are high and medical treatments are only available through medical institutions registered with the health insurance company.
In Japan, medical institutions submit health insurance claims data to health insurance unions to claim treatment expenses covered by insurance. One health insurance claim is created for each patient every month. Medical fees are defined precisely according to the medical services provided and also included in the health insurance claim. These enormous amount of health insurance claims, specific health checkups, specific health guidance data etc. of all residents are accumulated in the national database called NDB.
NDB was built to conduct surveys and analyses necessary for preparing, implementing, and evaluating a medical expenses optimization plan based on the “Act on Assurance of Medical Care for Elderly People” which was put in place in April 2008. However, there were several issues when it came to using the current NDB.
- Rigid system structure, a characteristic of relational databases (RDBs) One issue was the rigid system structure, which is a characteristic of RDB. In the future, nursing care services data and detailed health checkup data will also be stored in NDB, in addition to the conventional data which is increasing in recent years. However, it is difficult to change data formats of the data stored in RDBs since they require rigid data structure. Furthermore, time consuming and expensive physical server replacements are required to enhance the system performance. These factors made it difficult to flexibly cope with the increasing amount of data and data types.
-
Various rework required at each step of data use
Users such as researchers and analysts have also had problems because they had limited opportunities to look at the NDB data specifications and fewer opportunities to use NDB data. This is because, a facility that satisfies extremely high security requirements with computation resources to process enormous amounts of data is required to analyze NDB data. In general, researchers/analysts or institutions cannot afford to have such facilities. In order to solve this problem, the On-site Research Center of the NDB has been established in Kyoto University and The University of Tokyo.
However, researchers and analysts had to go all the way to the on-site research center even to understand how to analyze the data. Even if they go to the center, it took a great deal of time to obtain the desired result because it required a great deal of refactoring work involving trial and error. Therefore, users unfamiliar with the NDB system that has more than 10 billion records may not be able to obtain a result, even after several days of work depending on their data search and analysis method. -
Cumbersome data conversion work required to reorganize the data into structured data suitable for various researches
Another factor that significantly affected the convenience of using NDB data was that users had to carry out cumbersome preprocessing such as data reorganization and conversion on their own according to the contents of research and analysis.
An especially significant issue was the "ID problem", which prevented the continuous analysis when a patient's ID changed at some point in time. The NDB records do not contain patient names due to privacy concerns. Therefore, changes in patient IDs due to marriage, retirement, and other life events, as well as typographical errors prevent NDB users from tracing data at the time of analysis.
As a research project in FY 2016, the Japan Agency for Medical Research and Development (AMED, hereafter) solicited a research project called "Research for Infrastructure Construction of Studies with the Next Generation NDB Data to Create New Evidence" to solve these problems of current NDB.
Points of the selection
Essential requirements: Flexible scalability and avoidance of vendor lock-in
Members of the project included professionals familiar with both medical and informatics fields from Kyoto University Hospital, Nara Medical University, and Graduate School of Medicine and Faculty of Medicine of The University of Tokyo. NTT DATA was selected to build the platform because of their rich track record in Hadoop/Spark platform development.
Professor Tomohiro Kuroda at Kyoto University, the coordinator of the project, says, "Adequate scalability was a key requirement in the public tender. If the system specification can only be enhanced through gradual steps involving server replacement as it was before, the system cannot cope with future increase in data and new data such as nursing care services data. Therefore, scalability with the future in mind is essential."
Another crucial requirement in the public tender was “a framework without vendor lock-in.” “After confirming the feasibility of the next-generation NDB through this project, new tenders will be called to build the production environment. Depending on the technology of a specific vendor tend to bring in disadvantages as a national project. Those disadvantages include unfairness, higher costs caused by lack of price competition, and restrictions in future system modifications,” continued Professor Kuroda.
Thus, the next-generation NDB had to solve the issues of the current NDB while satisfying the requirements as a public tender—flexible scalability and vendor lock-in avoidance. Two open-source software packages, Hadoop and Spark were selected as the core software of the next-generation NDB. Hadoop is capable of accumulating and processing large-scale data sets with distributed computing, and Spark is capable of efficiently processing iterative tasks that use large-scale data. These open-source software are based on scale-out design, thus the system performance can easily be increased by only adding commodity servers without using expensive high-performance servers.
"We had no choice but to use Hadoop as the software framework in order to solve the issues while satisfying the requirements of the public tender. Thus, we looked for a company that has technical competence and know-how on Hadoop/Spark, and found NTT DATA in the center of the community. Their leading worldwide track record of Hadoop-related development and frequent information sharing from the heart of the community proves that NTT DATA understands the core of technology. Also, in addition to the fineness of their work, corporate strength was also an important factor as a national project. In the light of these considerations, we decided that NTT DATA is the best company to handle this project," explains Professor Kuroda about the reasons for selecting Hadoop/Spark and NTT DATA.

Professor Tomohiro Kuroda
Medical Informatics, Graduate School of Medicine, Kyoto University
General Manager of Division of Medical Information Technology and Administration Planning, Kyoto University Hospital
Future prospects
For further utilization and evolution of next-generation NDB
The next-generation NDB has some areas that can be improved while we received both positive and constructive feedback from users regarding improving the next-generation NDB.
"In addition to the fact that all residents are subscribed to the public health insurance system in Japan, prices are set finely. So, it is possible to know the details of the treatments by just looking at the health insurance claims data. You cannot find any other country in the world that has health insurance data in this volume and detail. However, unfortunately, there was no proper mechanism to take advantage of these data which was sleeping behind closed doors. Next-generation NDB is the system to make use of this ‘treasure’. Having established a baseline system on top of the Hadoop/Spark platform in this project, we can clearly see our next steps from here. One of them is to provide a catalog that guides users to understand where and what kind of data are available in the platform. It will attract more users. As a result of this project, we can now create data marts from various angles and expect to be used for various researches. However, having too many data marts to select from makes it difficult to use the system. So, we plan to summarize them in a catalog, and update it after evaluating the demand trends. We have already proposed a mechanism for creating a catalog, and it will come in to light in a new project," explained Professor Kuroda about the future prospects of the next-generation NDB.
Also, Professor Kuroda expressed his expectations regarding NTT DATA saying, “NTT DATA is one of the few important companies that can attract people to accomplish this kind of innovative IT project on the grounds of technical competence, credibility, and track record. I hope NTT DATA will continue to actively provide the government and academic societies with proposals that change the society as a whole.”
The baseline platform has been built for the practical use of the next-generation NDB. NTT DATA will continue to fully support the next-generation NDB—an important platform that helps medical services in Japan.

Tomohiro Kuroda
Kyoto University (right)
Toru Shimogaki
NTT DATA (left)
Contents of the project
A dream team of specialists in each field with high technical competence
This project was launched in January 2017 and carried out with the following roles:
- - Kyoto University: Establishment of an environment that enables learning, trials, and operations related to the research using NDB data.
- - The University of Tokyo: Creation of data marts for analysis. Especially focus on advanced utilization of next-generation NDB with AI in mind.
- - Nara Medical University: Creation of data marts for analysis. Especially focus on improving the accuracy of patient search (ID matching).
- - NTT DATA: Building the high performing and scalable platform using Hadoop/Spark, and other required software.
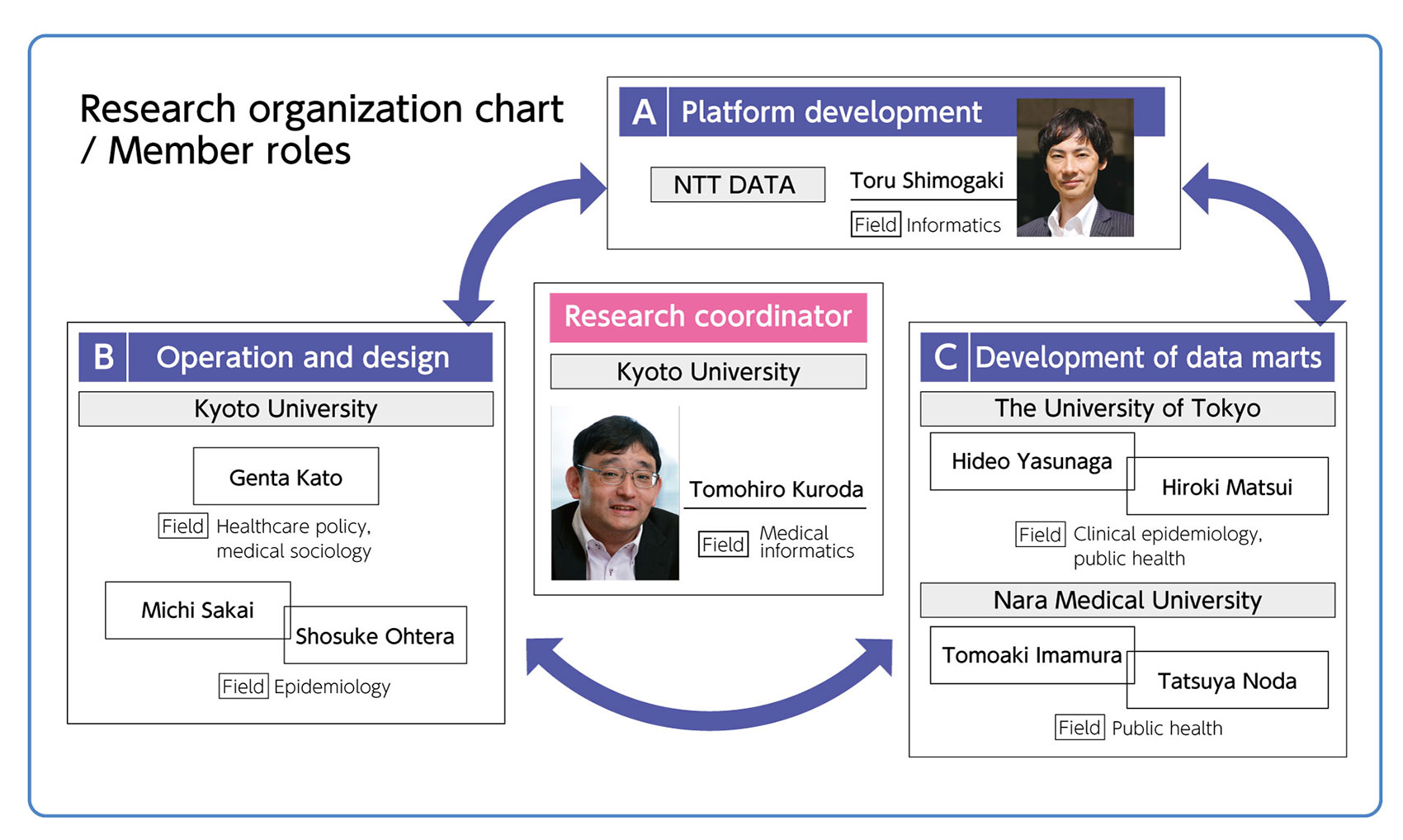
Professor Kuroda who specializes in medical informatics supervised this project. He is responsible for managing digital medical data, utilizing those data in hospital operation and management. He is also familiar with analyzing medical data such as health insurance claims data. His past achievements and experiences largely contributed to the success of this project.
Professor. Kuroda says, "Specialists of different fields shared their goals and progress statuses in time, and this helped me a lot to manage the project. It was literally a dream team of aces. Among everything, I was amazed by the high technical competence of NTT DATA on Hadoop and Spark, their knowledge in the medical field and tasks related to health insurance claims, and their detailed project management skills."
Benefits of utilizing Hadoop/Spark
Realization of data research platform that enables high performance and flexible analysis
-
Realization of high performance and scalability by utilizing Hadoop/Spark
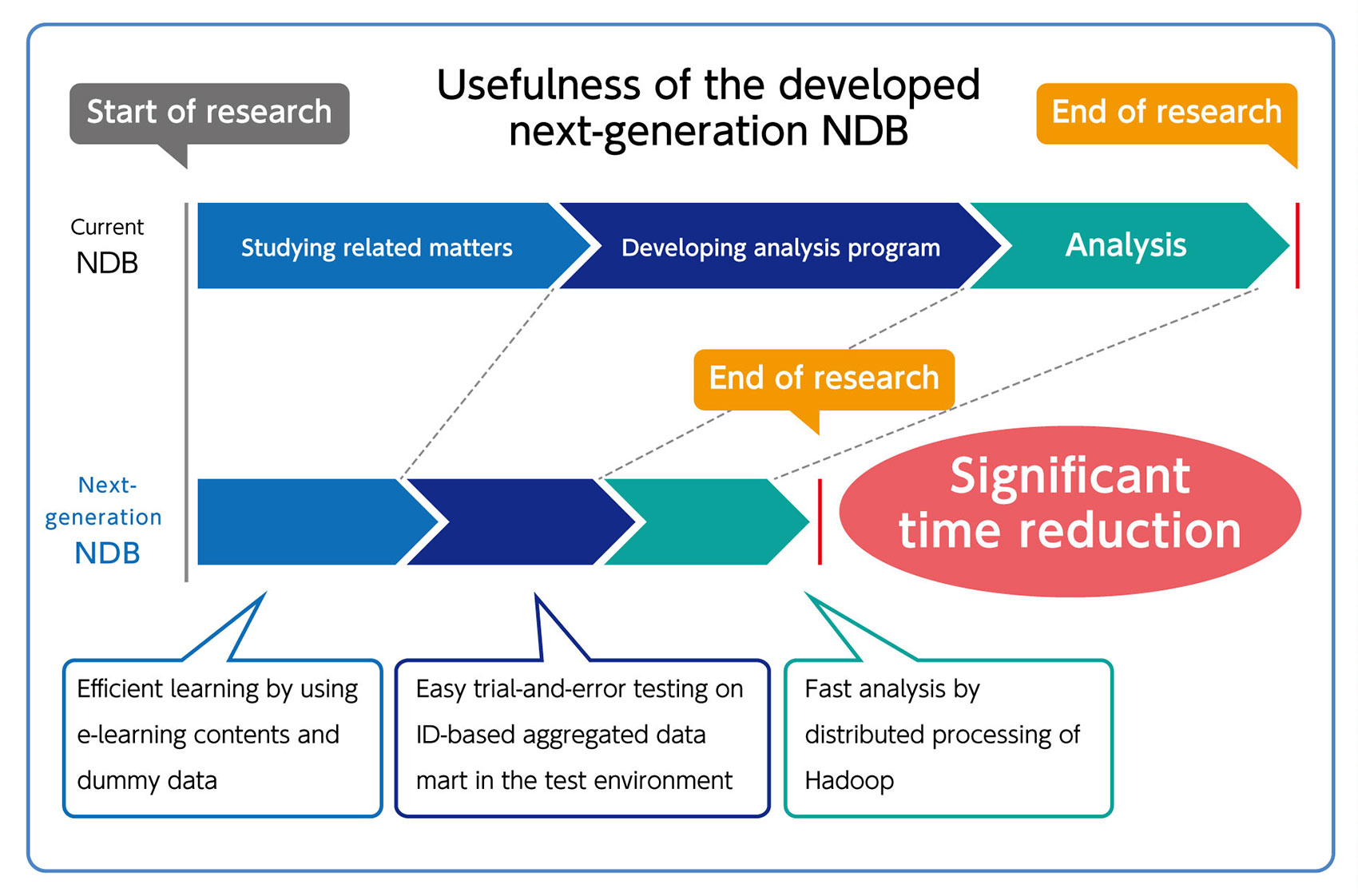
- Reduction of processing time from hours to minutes The project was started in January 2017. Construction of the next-generation NDB utilizing Hadoop/Spark was started from April 2017 after requirement definition phase. System tests using third-party data were started from November the same year, and data mart creation, analysis, and verification were carried out in January 2018. Data extraction, for example, which used to take four hours as an on-site job only takes several minutes in the next-generation NDB, thanks to the test environment and the preliminary data preparation. Researchers from different universities conducted analysis using the next-generation NDB and realized the effectiveness and speed of it.
In addition, the scalability of the Hadoop/Spark platform was verified and it was confirmed that the system performance can be increased by adding commodity servers. - Improvement of the learning efficiency by establishing "materials to learn the basics of health insurance claims" and "a learning environment where dummy data is available" Kyoto University prepared e-learning contents to learn the basics of health insurance claims for users who are not familiar with health insurance claims data and procedures. Kyoto University also prepared an NDB hands-on environment where users can search and do analysis using NDB dummy data, easily. With these enhancements, users can now efficiently learn and understand the basics of health insurance claims data by using the e-learning contents and by testing the analysis logic in the NDB hands-on environment. This helped beginners of NDB analysis to reduce the lead time until the actual analysis is started.
-
Increase of the analysis efficiency and convenience by preparing data marts required for analysis
The "ID problem" was particularly a critical issue among other issues such as data reorganization and data conversion when analyzing NDB data. NTT DATA successfully implemented the ID matching algorithm provided by Nara Medical University on the distributed platform. As a result, obstacles that blocked continuous data tracing were cleared, increasing the accuracy of patient ID-based data aggregation.
NTT DATA also implemented the analysis algorithms provided by The University of Tokyo, and prepared frequently used data marts in advance. As a result, research and analysis activities can now be performed efficiently and conveniently using these existing data marts without creating new data marts.
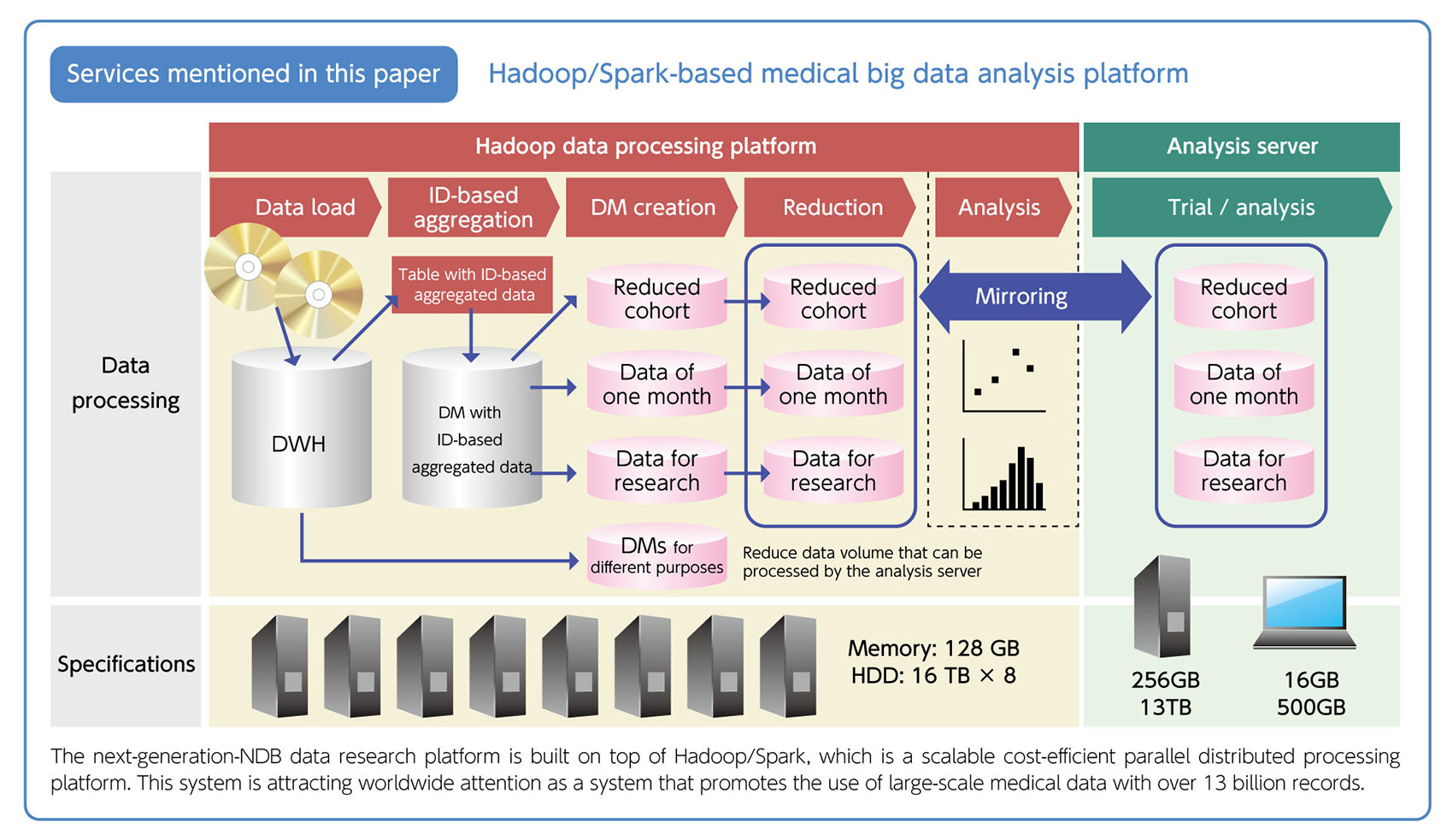
(Service mentioned in this paper)
- Hadoop/Spark-based medical big data analysis platform
The next-generation-NDB data research platform is built on top of Hadoop/Spark, which is a scalable cost-efficient parallel distributed processing platform. This system is attracting worldwide attention as a system that promotes the use of large-scale medical data with over 13 billion records.
Profile

- Company name
-
Japan Agency for Medical Research and Development (AMED)
- Location
-
20th floor of Yomiuri Shimbun Building, Otemachi 1-7-1, Chiyoda-ku, Tokyo, Japan
- Established on
-
April 1, 2015
- overview
-
Established with the purpose of conducting consistent research and development in the medical field from basic research to practical application using the capabilities of universities and research institutions. The agency is under the control of the Ministry of Education, Culture, Sports, Science and Technology; the Ministry of Health, Labour and Welfare; the Ministry of Economy, Trade and Industry; and the Cabinet Office.
