
Search and Extract data from RWE medical papers with Almirall
IGNACIO BARAHONA
Product Owner Dolffia and Generative AI Office Lead
Over 10 years of applied consulting experience as an industry leader. Currently serving as a manager on the Data & Analytics executive team at NTT DATA, he is entrusted with leading the Generative AI office, where he spearheads the development of cutting-edge solutions at the intersection of data and artificial intelligence.
Objective
This project focused on constructing an NLP-based solution that allows researchers to extract information from RWE medical papers in different formats in which they tested the effect of an active principle in patients with atopic dermatitis. The aim of this project was to reduce the time for extracting information from different data sources by creating an easily actionable knowledge database. This was done in two ways. First, generating an excel file with the answers that the researchers want to extract. Secondly, creating a Web App where they can do their own searches in the RWE medical papers.
Business problem
Researchers can spend many hours manually reviewing papers related to relevant drug and extracting information from them. Being able to extract the information from papers automatically, and to easily find papers with specific characteristics would help them reduce time invested into analyzing how science and studies are evolving. In consequence, this would reduce the cost of the task while giving them a tool that can help them work more efficiently.
Methods and solution
The approach that we followed consisted of developing a pipeline for preprocessing papers, then indexing the information and finally developing a Q&A system that can retrieve the indexed information and form an exact answer. The full diagram of the solution is shown in Figure 1.
Preprocessing pipeline
To extract information for the papers, they previously need to be preprocessed in a specific manner. This not only includes extracting the text with Optical Character Recognition (OCR) techniques, but also the extraction of tables independently and segmentation of the paper into sections. To facilitate the segmentation of the paper, we needed to find titles of the different sections. This title identification is dependent on text characteristics such as font, font details (bold, italics), font size, etc.
Therefore, the first step in the preprocessing pipeline if the text extraction, along with their characteristics. This includes extracting coordinates of each word (that is, the location of the word in the pdf page) that becomes relevant when removing information or clustering words into paragraphs. In parallel, tables are extracted and processed, which is one of the most complex operations of the pipeline. Firstly, because the table can be in any format (one or two columns, in between text, be a full page, etc.). Secondly, it needs to be extracted, processed (including merged cells, shared titles, different sections within the table, different number of columns and rows, etc.) and saved in a format that could be indexed and understood by our information extraction pipeline. Finally, the text included in a table must be deleted from the main text to avoid getting noise from the information of the table (that is usually numbers). This considers that tables normally occur in the middle of the text and sometimes they occupy both columns of an article (when it is a two-column paper).
The cleaned extracted text is then processed into paragraphs so that valuable information is not divided when text is indexed as individual chunks and so the retrieval is able to understand the context better. Here it is also where the table deletion becomes very relevant. Using the text characteristics previously extracted, we can determine whether a line/paragraph is a title or not. Titles are then used to identify sections (such as Introduction, Methodology, Results, References…) and removed from the cleaned extracted text. Article titles are also identified to divide articles in case there is more than one paper within a single document. Following this, some sections can be removed (such as the Reference section) as it contains too much noise for the Information Extraction pipeline.
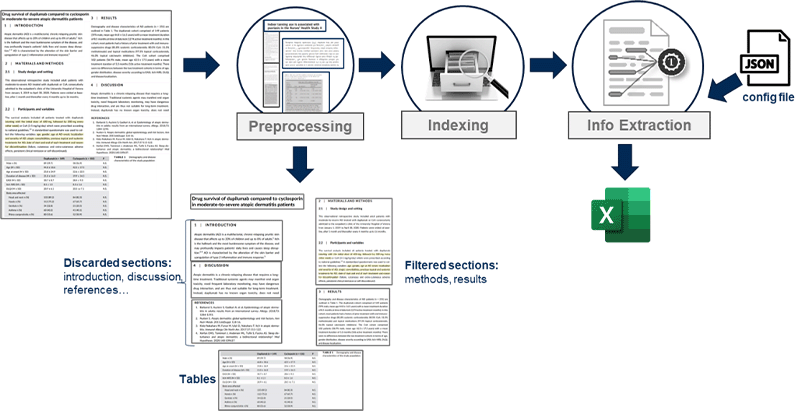
Figure 1. Solution diagram. Papers are firstly preprocessed (text and table extraction, segmentation into sections, clean-up of the text, paragraph creation) and indexed. Then information is extracted, using a config JSON file containing the details that need to be extracted, and outputted into a CSV file.
Information extraction pipeline
After preprocessing the text, both paragraphs and tables are indexed in Elastic (ref), alongside metadata with the section (Introduction, Methods, Results, Discussion…), whether it is text or table and the name of the paper. These metadata help the retrieval and filtering find the best results. To retrieve the documents, we have used a combination of BM25 and dense passage retrieval (DPR) open-source models. Combining both models give us an advantage for any kind of query input, since BM25 tends to work better with keywords and DPR is better with the semantic meaning of sentences.
When a question is asked, documents (meaning, chunks of text or tables) are retrieved and the top 3 answers are sent to the Q&A system alongside the question. The Q&A system that we have developed builds a prompt with said question and the retrieved document. This prompt highly depends on the type of question (i.e., subpopulation effects, percentages of adverse effects, scores…) that is asked. Besides, different questions can prioritize the retrieval of a paragraph or a table (for example, percentages tend to appear in tables) and thus facilitate the correct answer. The created prompt is then sent to GPPT-3 via API, using the davinci-003 model. The answer that GPT-3 gives is then processed and returned to the user.
Excel generation
The JSON config file contains all questions that need to be responded, their type and all needed details to form the prompt. This JSON config file is required to generate the output excel file. When the process is launched, using the JSON file, the system goes through each one of the questions, retrieves the documents, creates the prompt, sends it to GPT-3, gets the answer and saves it in an excel file. It performs these operations for each one of the papers included. The final excel contains the answers for each paper and each question. These answers can either be a written answer (like a percentage) or a cell colored in Green if the response is True (or if a given concept/score appears) or not colored if it does not appear in the paper.
WebApp
The website created can be found in the following URL: almirall.dolffia.com/. This website contains two pages. The main page where searches can be done by a user and a back-office for adding papers and generating the excel. Searches done in the main page can be filtered by paper name and type (table or text) and can be either a question such as <What is the % of patients with conjunctivitis as adverse effect?> or keywords such as <alopetia areata>. Besides, the search engine allows for an exact search done by adding quotes. That is, <Can the treatment improve "itch" in patients?> will only return fragments that contain the word "itch". This search will be done across papers previously uploaded and indexed in the back-office page.In the back-office page, a user can upload new papers, delete them, and index them. Once they have been processed, they can be chosen to run through the Information Extraction pipeline. To do so, the user must include a configuration JSON file with the questions and details and select the papers that they want to extract. After launching the pipeline, the excel file containing all the answers can be easily downloaded from this page.
Results
Extracted concepts are included in an excel file, achieving more than 88 % accuracy.
In this project, we focused on answering 29 concepts provided by a pharmaceutical company from RWE medical papers. Seven of those consisted of extracting a percentage of patients (or an empty field if not found) with either concomitant symptoms or adverse events. For example: "What is the percentage of patients that developed asthma as an adverse effect?". The other 22 consisted of returning whether a concept is included in the paper or not (Yes/No questions). The latter included concepts such as subpopulation effects (efficacy differences), and scores used to measure general efficacy, specific symptoms, quality of life and wellbeing and general health PROS.
These questions were answered using the system developed here for a total of 80 RWE medical papers. These papers were preprocessed, indexed and, with the JSON config file, ran through the extraction pipeline to generate the final excel.
To evaluate our pipeline, the results returned by our system were compared to the ones manually extracted by the client. Final metrics, including both the accuracy and the f1-score, are shown in Figure 2. As it is shown, we achieved a high accuracy in most fields, with an average of 88.6 ± 11.5 % accuracy. Concepts are classified into four categories: simple scores, complex scores, percentage extraction (concomitant symptoms and adverse effects), and segmentation characteristics (subpopulation effects). Accuracies are higher than 90 for all simple scores (97 ± 3 %) and complex scores (96 ± 0.8 %), while questions for percentage extraction have a mean accuracy of 81.2 ± 8 %. Questions of segmentation characteristics are more variable (79.88 ± 15.9 %). This is because the effect of a parameter (sex, age, BMI, etc.) in the efficacy of a drug is not always very well defined in the papers. Our system is trained to look for statistical significance of the effect (that is, a p-value < 0.05) and this is not always specified in papers. Sometimes they divide the study population into subgroups based on one of these parameters and show different values, but do not include a statistical analysis to verify differences are significant. Other times, they show the difference in a visual graph that our system cannot yet comprehend. Another parameter that has a low accuracy is "bionaïve", which defines a patient that has not received a treatment in the past. For this parameter, we need to extract whether patients are bionaïve or not. The main problem to do so, is that it is not always specified and thus we must make assumptions. Other times, only a group of them were not bionaïve and they went through a wash-up period before the study, which can be confusing for our system. The accuracy of these concepts could be improved in the future by shortening the scope of the questions and dividing them in several. For example, considering whether there is a single patient that has tried the treatment before, or whether sex or age is considered when measuring efficacy (without considering the statistical significance) and if so, then checking if a statistical analysis was done, and if so, whether it was significant.
Overall, the system could return most questions with a high accuracy, finding the answer even in cases where a user might miss it. In fact, we detected more than 50 answers in which our system out-performed human extraction.
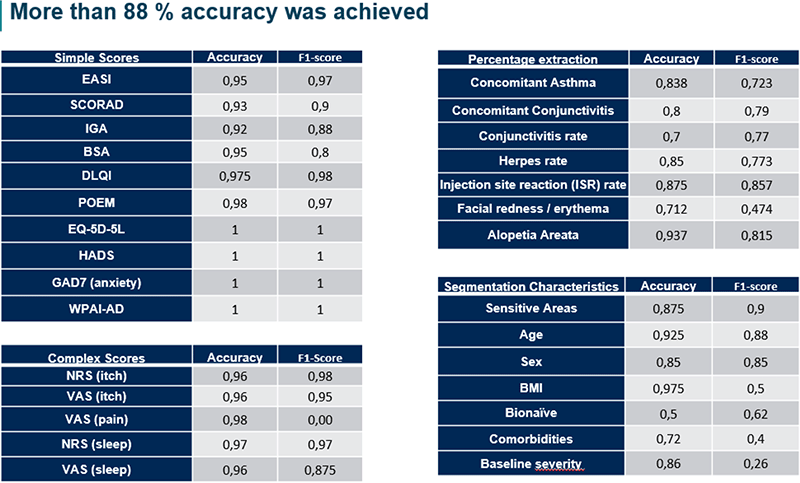
Figure 2.
Web App.
The designed WebApp has a clear interface with a user-friendly back-office page to upload papers, extract the information from it and download the excel files shown in the previous section (Figure 3). Moreover, we included the search engine so a user can ask questions to the uploaded documents, being able to filter by paper, type (text/table) and limit the number of results.
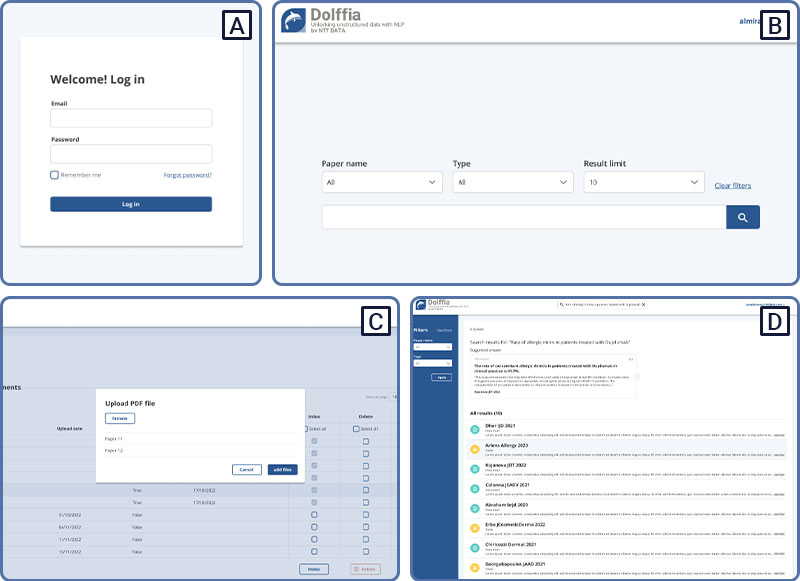
Figure 3. Web App. (A) Login page. (B) Search engine, with functionalities to filter by paper, type (text or table) and change the number of results shown). (C) Backoffice for uploading and processing documents.
Voice of the customer
"The medical team required to extract scientific information that is spread along many external publications. This solution enables the automation of searching and filtering for all relevant articles and extracting the valuable information. We save a lot of time (depending on the type of analysis needed) and ensure that our medical experts devotes their time to what is most valuable".
Gonzalo Durbán Diez de la Cortina, Head of Data & Analytics at Almirall
Conclusions
Here, we have developed a system that can help researchers extract information from papers efficiently. With accurate results and low processing-times, this system can answer previously defined questions from a set of RWE medical papers. It is also scalable, since it allows to include a high number of papers that could not be processed manually. Besides, the search engine allows to ask new questions in papers that can be easily uploaded thanks to the website.
Therefore, we believe the pipeline that we have designed can help researchers reduce time in extracting information and improve accuracy by eliminating human errors. This pipeline can also be expanded to new scopes (both new drugs for the same field and different fields, or even add new questions) by doing tweaking the JSON configuration file. This makes our system highly adaptable.
